Researchers reveal the universal mechanisms of DNA and RNA deformations
DNA and RNA, the two main types of nucleic acid and the building blocks of life, are susceptible to environmental stimuli, which can cause them to deform, bend or twist. These deformations can significantly affect gene regulation and protein functions, but they are extremely difficult to measure using traditional techniques. Recently, a research team co-led by a physicist from City University of Hong Kong (CityU) accurately measured the change in a nucleic acid induced by salt, temperature change and stretching force. Their findings help reveal the underlying universal deformation mechanisms of DNA and RNA.
While DNA and RNA deformations are of great biological importance, our understanding of them is limited due to the challenge of making precise measurements of nucleic acid deformations and the complexity of nucleic acid interactions. To overcome these two difficulties, a research team led by scientists from CityU and Wuhan University used a combination of experiments, simulations and theories to investigate the universality of DNA and RNA deformations.
The success of the research lies in an accurate measuring tool, called magnetic tweezers (MT). This is a powerful experimental technique used in biophysics and molecular biology to study the mechanical properties of biological molecules, such as DNA, RNA and proteins. In a magnetic tweezers experiment, a tiny magnetic bead is attached to a molecule of interest, and a magnetic field is applied to manipulate the position of the bead.
By measuring the movement of the bead, researchers can study the mechanical properties of the molecule, such as its elasticity, stiffness, and response to external force. This can be used to measure tiny DNA and RNA twist changes caused by environmental stimuli. Even small twist changes can accumulate along a long DNA or RNA molecule and cause a large rotation of the DNA or RNA end.
In the experiments, the team used magnetic tweezers to precisely measure DNA and RNA twist changes induced by salt, temperature change, and stretching.
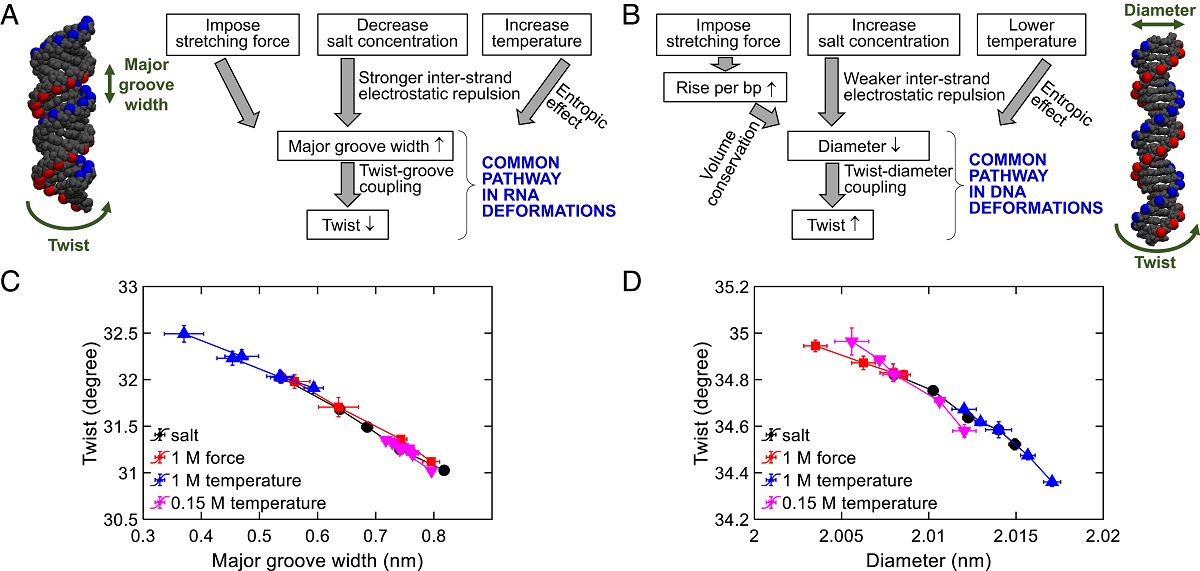
Through the experiments, the team quantified the DNA twist-diameter coupling constant and the RNA twist-groove coupling constant and applied the coupling constants to explain DNA and RNA deformations. By combining these findings with simulations, theory and other previous research results, the team found that the DNA and RNA deformation mechanisms induced by salt, temperature change and stretching force are driven by two common pathways: twist-diameter coupling for DNA and twist-groove coupling for RNA.
For DNA, environment stimuli usually modify the diameter of the DNA first, and then cause a twist change through a strong coupling between DNA twist and diameter. But for RNA, lowering the salt concentration, or increasing the temperature “unwinds” the RNA because this enlarges RNA major groove width and causes a decrease in the twist. Hence, this is called twist-groove coupling.
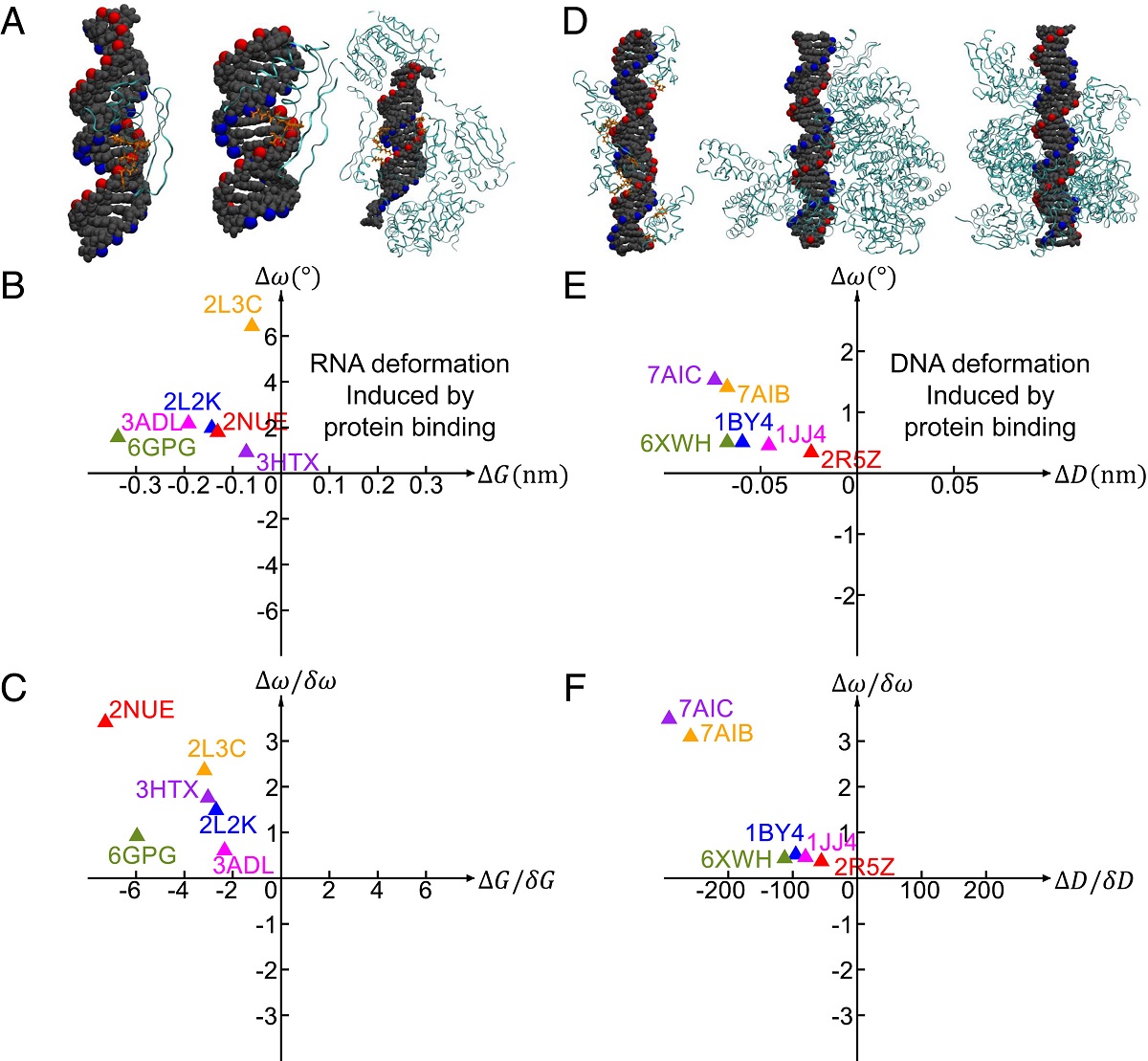
By analyzing data from other studies on protein binding, the team found that the DNA and RNA follow the same common pathways when deformation is induced by protein binding, suggesting that the two pathways are utilized to reduce the DNA and RNA deformation energy cost to facilitate protein binding.
Their findings suggest that the physical principles underlying nucleic acid deformation are universal and can be applied to different types of nucleic acids and environmental stimuli.
“The latest findings can be applied to better understand DNA packaging in cells and the related deformation energy cost. The results also provide insights into how proteins recognize DNA and RNA and induce deformations, which are the key steps in gene expression and regulation,” said Professor Dai Liang, Associate Professor in the Department of Physics at CityU, who co-led the research.
The results were published in the scientific journal Proceedings of the National Academy of Sciences (PNAS) under the title “Universality in RNA and DNA Deformations Induced by Salt, Temperature Change, Stretching Force, and Protein Binding”.

The first authors are Tian Fujia, from CityU, and Zhang Chen and Zhou Erchi, from Wuhan University. The corresponding authors are Professor Dai, from CityU, and Professor Zhang Xinghua, from Wuhan University. Key funding sources are the Research Grants Council of Hong Kong and the National Natural Science Foundation of China.