Novel tool targeting unusual RNA structures for potential therapeutic applications
Ribonucleic acids (RNAs), which decode the genetic code stored in DNA and produce proteins, fold into diverse structures to govern fundamental biological processes in all life forms, including humans. Targeting disease-associated RNA structures with drug-like small molecules has been one of the gold standards for developing RNA-targeting drugs in the scientific field. Recently, a research team at City University of Hong Kong (CityU) developed a new type of RNA structure targeting tool to specifically recognise unusual four-strand RNA structures, which are associated with diseases such as cancer and neurological disorders. The findings could help develop new therapeutic tools for related treatments.

The research was led by Dr Kwok Chun-kit, Associate Professor in the Department of Chemistry and the State Key Laboratory of Marine Pollution at CityU. It was published in Nature Protocols, titled “Development of RNA G-quadruplex (rG4) targeting L-RNA aptamers by rG4-SELEX”.
RNA G-quadruplexes: linked to cancer and neurological disorders
“We are the first group in the world to develop “rG4-targeting L-RNA aptamers”, a new type of tool that can target RNA G-quadruplex (rG4). Our data suggests that these newly developed L-RNA aptamers have exquisite binding affinity and selectivity towards rG4 targets, and often outperformed the existing state-of-the-art rG4-targeting small molecules,” said Dr Kwok. He described their achievements as “encouraging” and added “it offers new and important directions into the selective targeting of rG4s, and potentially to other unusual nucleic acid structures in DNA and RNA as well.”
What is rG4? It is a special structure formed by RNA sequences that can be found in humans, plants, bacteria and viruses. It can be a structural scaffold for protein binding and can perform different biological functions. Individual rG4 candidates have been found to have important functional and gene regulatory roles in cells. Recent studies have also linked rG4s to diseases such as cancer, neurological disorders and viral pathogenesis.
To further study rG4s, the first step is to locate them. Over the years, the major approach to targeting functional rG4s is based on small molecule ligands, meaning organic or inorganic compounds with low molecular weight. “However, most of the rG4 ligands developed cannot differentiate rG4s with similar structures, posing a significant challenge in achieving selective targeting of specific rG4s for gene control and manipulation,” explained Dr Kwok.
Targeting rG4s with novel L-RNA aptamers
To overcome this challenge, Dr Kwok and his team developed “L-RNA aptamer-based rG4 targeting”, a brand-new solution to this longstanding problem. Aptamers are short, single-stranded DNA or RNA molecules that fold into defined structures and bind to specific targets, such as metal ions and proteins.
Dr Kwok and his team have spent almost two years establishing an aptamer-selection platform, called “rG4-SELEX”, to develop different L-RNA aptamers that target rG4 of interest.
In their experiment, the team discovered that the L-RNA aptamer they call “L-Apt.3-7” bound to its intended rG4 target tightly and did not mistake it for other similar rG4 structures. This highlighted that selective rG4 targeting can be achieved using an L-RNA aptamer, a new discovery.
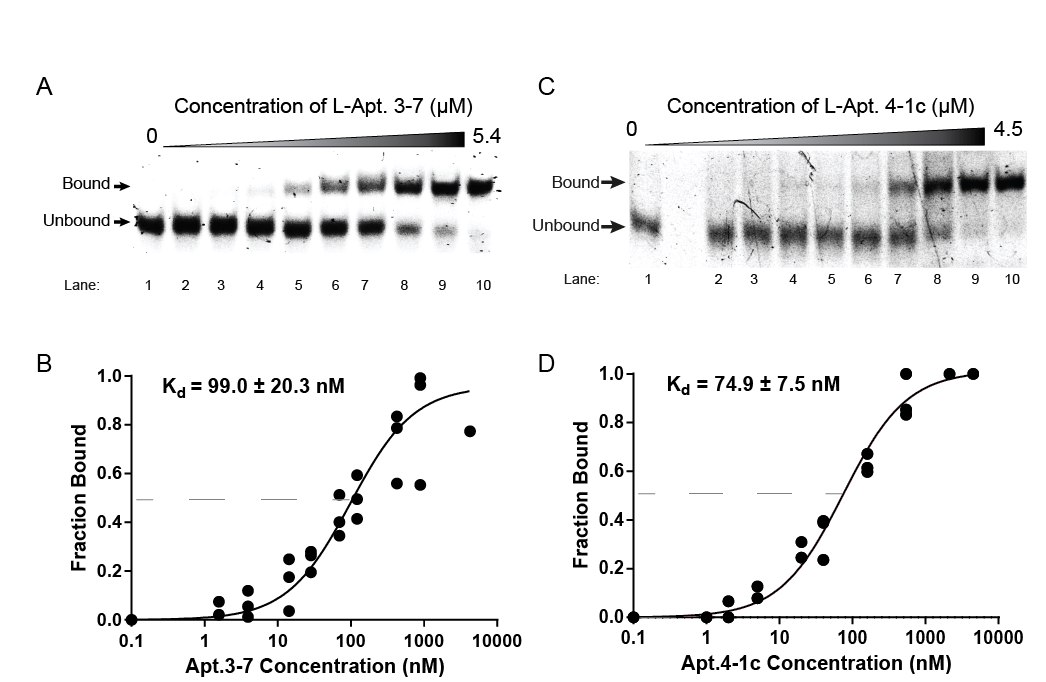
(Photo source: Mubarak I. Ishaq, Chun-Yin Chan et al./DOI number: 10.1038/s41596-022-00679-6)
The team also demonstrated that L-RNA aptamers can be utilised to interfere with rG4-protein interactions to stop their biochemical and cellular functions, which can potentially be manipulated for therapeutic purposes. Their experimental results showed that L-RNA aptamers’ performance in the rG4-protein interference is at least equal to, if not better than, the state-of-the-art rG4-targeting ligands.
Uncharted territory with limitless potential
“In this research, we introduced the first universal framework for the robust development of L-RNA aptamers for targeting biologically and therapeutically relevant rG4 structures and rG4-protein interactions. Our findings offer a new targeting strategy for potential disease treatment in the future,” explained Dr Kwok.
Dr Kwok and his team are currently conducting research on applying rG4-targeting L-RNA aptamers to target a number of disease-related rG4 structures and rG4-protein interactions in coding and decoding human RNA.
Dr Kwok anticipated that further refinements of this aptamer-selection platform would enable diverse RNA structure-targeting L-aptamers to be developed in a high-throughput manner, which can potentially be applied to combat human diseases, such as COVID-19. “It is my strong belief that pursuing technological innovation will naturally drive our scientific exploration and deeper understanding of biology,” he added.

Dr Kwok is the corresponding author of the paper. Dr Umar Mubarak Ishaq and Mr Chan Chun-yin, both previously from the Department of Chemistry and the State Key Laboratory of Marine Pollution at CityU, are the first authors of the paper.
The work was supported by CityU, the Research Grants Council in Hong Kong, the Croucher Foundation, the Shenzhen Basic Research Project, and CityU’s State Key Laboratory of Marine Pollution.
DOI number: 10.1038/s41596-022-00679-6